Diabetes Prediction Using Machine LearningDiabetes is a medical disorder that impacts how well our body uses food as fuel. Most food we eat daily is converted to sugar, commonly known as glucose, and then discharged into the bloodstream. Our pancreas releases insulin when the blood sugar levels rise. Diabetes can cause blood sugar levels to rise if it is not continuously and carefully managed, which raises the chance of severe side effects like heart attack and stroke. We, therefore, choose to forecast using Python machine learning. Steps
Installing the LibrariesWe first have to import the most popular Python libraries, which we will use for implementing machine learning algorithms in the first step of building the project, including Pandas, Seaborn, Matplotlib, and others. We will use Python because it is the most adaptable and powerful programming language for data analysis purposes. In the world of software development, we also use Python. Code The Sklearn toolkit is incredibly practical and helpful and has practical applications. It offers a vast selection of ML models and algorithms. Importing the DatasetWe are using the Diabetes Dataset from Kaggle for this study. The National Institute of Diabetes and Digestive and Kidney Diseases is the original source of this database. Code Output <class 'pandas.core.frame.DataFrame'> RangeIndex: 768 entries, 0 to 767 Data columns (total 9 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 Pregnancies 768 non-null int64 1 Glucose 768 non-null int64 2 BloodPressure 768 non-null int64 3 SkinThickness 768 non-null int64 4 Insulin 768 non-null int64 5 BMI 768 non-null float64 6 DiabetesPedigreeFunction 768 non-null float64 7 Age 768 non-null int64 8 Outcome 768 non-null int64 dtypes: float64(2), int64(7) memory usage: 54.1 KB As we can see, all of the columns are integers, except for the BMI and DiabetesPedigreeFunction. The target variable is the labels with values of 1 and 0. A person's diabetes status is indicated by a one or a zero. Code Output
Filling the Missing ValuesThe next step is cleaning the dataset, which is a crucial step in data analysis. When modelling and making predictions, missing data can result in incorrect results. Code Output Pregnancies 0 Glucose 0 BloodPressure 0 SkinThickness 0 Insulin 0 BMI 0 DiabetesPedigreeFunction 0 Age 0 Outcome 0 dtype: int64 We found no missing values in the dataset, yet independent features like skin thickness, insulin, blood pressure, ;and glucose each have some 0 values, which is practically impossible. A particular column's mean or median scores must be used to replace unwanted 0 values. Code Output
Let's now examine the data statistics. Code Output
Now our dataset is free of missing and unwanted values. Exploratory Data AnalysisWe will demonstrate analytics using the Seaborn GUI in this tutorial. Correlation Correlation is the relationship between two or more variables. Finding the important features and cleaning the dataset before we begin modelling also helps make the model efficient. Code Output 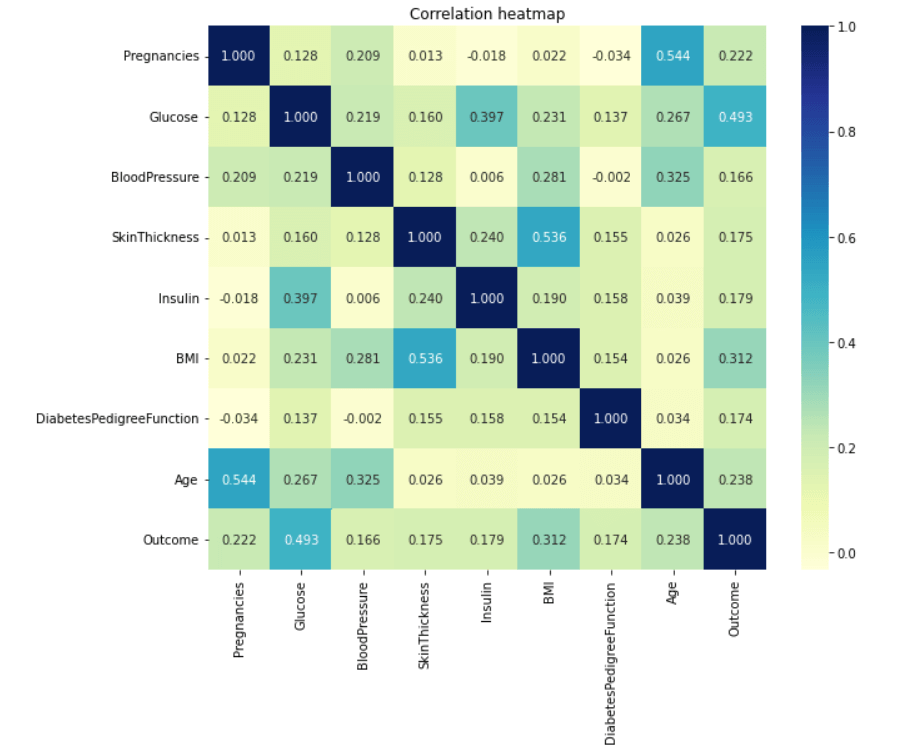 Observations show that characteristics like pregnancy, glucose, BMI, and age are more closely associated with outcomes. I demonstrated a detailed illustration of these aspects in the following phases. Pregnancy Code Output 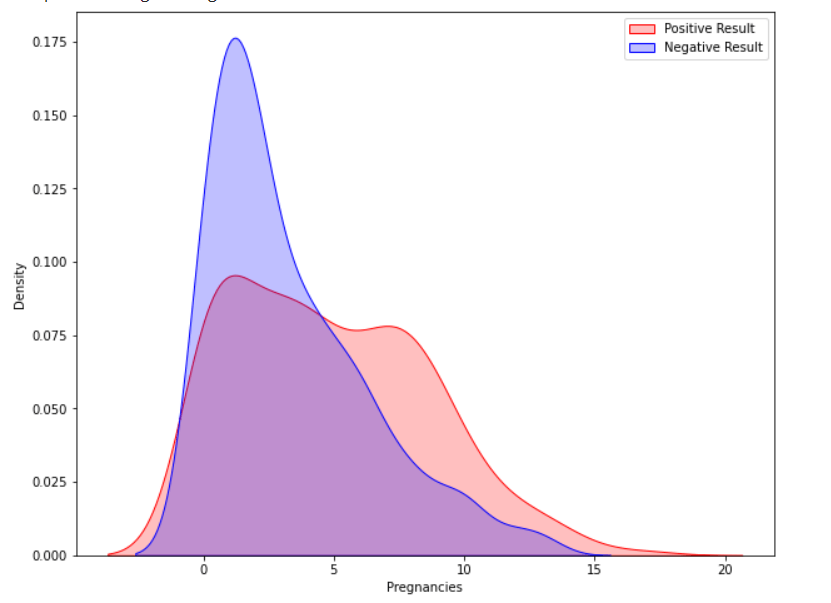 According to the data, women having diabetes have given birth to healthy infants. However, the risk for future complications can be decreased by managing diabetes. The risk of pregnancy issues, such as hypertension, depression, preterm birth, birth abnormalities, and pregnancy loss, is increased if women have uncontrolled diabetes. Glucose Output 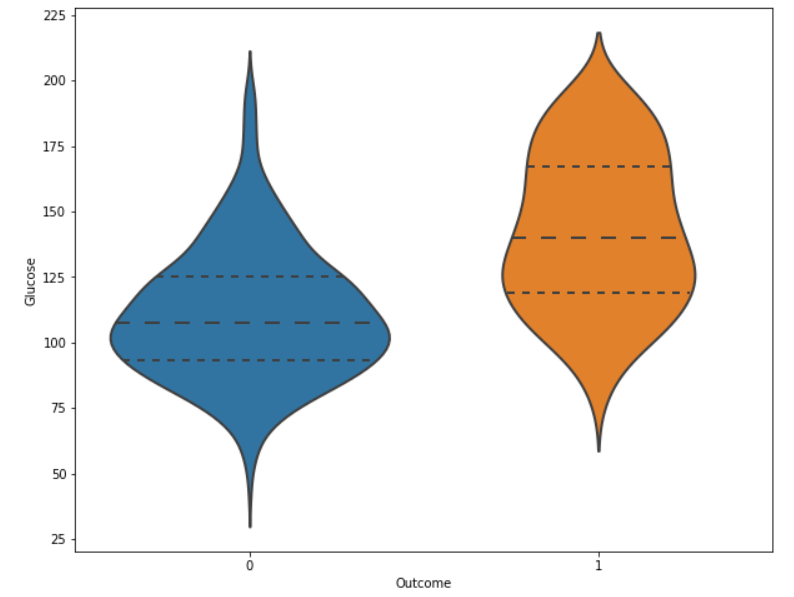 The likelihood of developing diabetes gradually climbs with glucose levels. Code Output 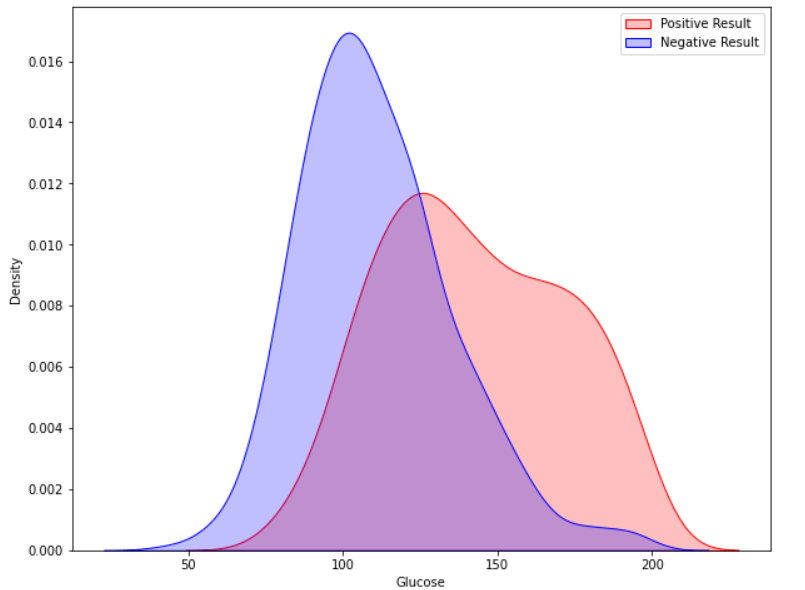 Implementing Machine Learning ModelsWe will test many machine learning models and compare their accuracy in this part. After that, we will tune the hyperparameters on models with good precision. We will use sklearn.preprocessing to convert the data into quantiles before dividing the dataset. Code Output
Data Splitting We will now divide the data into a training and testing dataset. We will use the training and testing datasets to train and evaluate different models. We will also perform cross-validation for multiple models before predicting the testing data. Code Output The size of the training dataset: 3680 The size of the testing dataset: 2464 The above code splits the dataset into the train (70%) and test (30%) datasets. Cross Validate Models We will perform cross-validation of the models. Code A list of machine learning models is passed to the 'cv_model' function, which provides a graph of the cross-validation scores based on the mean of the accuracy values of various models supplied to the function. Code Output
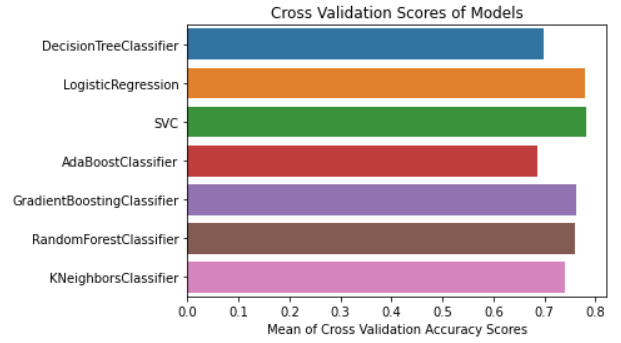 According to the above analysis, we have discovered that the RandomForestClassifier, LogisticRegression, and SVC models have higher accuracy. We will therefore perform hyperparameter tuning on these three different models. Hyperparameter Tuning Selecting the best collection of hyperparameters for a machine learning algorithm is known as hyperparameter tuning. A model input called a hyperparameter has its value predetermined before the learning phase even starts. Hyperparameter tuning is essential for machine learning models to work. We have individually tuned the RandomForestClassifier, LogisticRegression, and SVC models. Code GridSearchCV and the classification report classes are firstly imported from the Sklearn package. The "analyse grid" method, which will display the predicted result, is then defined. We have invoked this method for each model we utilised in SearchCV. We will tune each model in the following stage. Tuning Hyperparameters of LogisticRegression Code Output Tuned hyperparameters: {'C': 200, 'penalty': 'l2', 'solver': 'liblinear'}
Accuracy Score: 0.7715000000000001
Mean: 0.7715000000000001, Std: 0.16556796187668676 * 2, Params: {'C': 200, 'penalty': 'l2', 'solver': 'liblinear'}
The classification Report:
Mean: 0.7715000000000001, Std: 0.16556796187668676 * 2, Params: {'C': 100, 'penalty': 'l2', 'solver': 'liblinear'}
The classification Report:
Mean: 0.7675, Std: 0.16961353129983467 * 2, Params: {'C': 10, 'penalty': 'l2', 'solver': 'liblinear'}
The classification Report:
Mean: 0.7675, Std: 0.17224619008848932 * 2, Params: {'C': 1.0, 'penalty': 'l2', 'solver': 'liblinear'}
The classification Report:
Mean: 0.711, Std: 0.1888888562091475 * 2, Params: {'C': 0.01, 'penalty': 'l2', 'solver': 'liblinear'}
The classification Report:
precision recall f1-score support
0 0.78 0.88 0.83 201
1 0.70 0.53 0.61 107
accuracy 0.76 308
macro avg 0.74 0.71 0.72 308
weighted avg 0.75 0.76 0.75 308
As we can see in the output, the best score returned by the LogisticRegression model is 0.77 with {'C': 200, 'penalty': 'l2', 'solver': 'liblinear'} parameters. Similarly, we will perform parameter tuning for other models. Tuning Hyperparameters of SVC Code Output Tuned hyperparameters: {'C': 1.0, 'gamma': 0.0001, 'kernel': 'rbf'}
Accuracy Score: 0.7695158871629459
Mean: 0.745607333842628, Std: 0.019766615171568313 * 2, Params: {'C': 200, 'gamma': 0.0001, 'kernel': 'rbf'}
The classification Report:
Mean: 0.7521291344820756, Std: 0.02368565638376449 * 2, Params: {'C': 100, 'gamma': 0.0001, 'kernel': 'rbf'}
The classification Report:
Mean: 0.7542370483546955, Std: 0.046474062764375476 * 2, Params: {'C': 10, 'gamma': 0.0001, 'kernel': 'rbf'}
The classification Report:
Mean: 0.7695158871629459, Std: 0.016045599935252022 * 2, Params: {'C': 1.0, 'gamma': 0.0001, 'kernel': 'rbf'}
The classification Report:
Mean: 0.650001414707297, Std: 0.002707677330225552 * 2, Params: {'C': 0.01, 'gamma': 0.0001, 'kernel': 'rbf'}
The classification Report:
precision recall f1-score support
0 0.74 0.88 0.80 201
1 0.64 0.42 0.51 107
accuracy 0.72 308
macro avg 0.69 0.65 0.66 308
weighted avg 0.71 0.72 0.70 308
SVC Model's maximum accuracy is 0.769, somewhat less than that of Logistic Regression. We can leave this model here only. Tuning Hyperparameters of RandomForestClassifier Code Output Tuned hyperparameters: {'criterion': 'entropy', 'max_depth': 5, 'max_features': 'log2', 'n_estimators': 500}
Accuracy Score: 0.7717369776193306
Mean: 0.7673938262173556, Std: 0.0027915297477680364 * 2, Params: {'criterion': 'entropy', 'max_depth': 4, 'max_features': 'log2', 'n_estimators': 500}
The classification Report:
Mean: 0.7717369776193306, Std: 0.005382324516419591 * 2, Params: {'criterion': 'entropy', 'max_depth': 5, 'max_features': 'log2', 'n_estimators': 500}
The classification Report:
Mean: 0.7652151769798828, Std: 0.02135846347536185 * 2, Params: {'criterion': 'entropy', 'max_depth': 6, 'max_features': 'log2', 'n_estimators': 500}
The classification Report:
precision recall f1-score support
0 0.76 0.87 0.81 201
1 0.66 0.50 0.57 107
accuracy 0.74 308
macro avg 0.71 0.68 0.69 308
weighted avg 0.73 0.74 0.73 308
Predicting Unseen DataWe have spent time working on the Exploratory Data Analysis, cross-validation of the machine learning algorithms, and hyperparameter tuning to identify the best model that fits my dataset. We will now make predictions using the model of tuned hyperparameters with the highest accuracy score. Code Output precision recall f1-score support
0 0.78 0.88 0.83 201
1 0.70 0.53 0.61 107
accuracy 0.76 308
macro avg 0.74 0.71 0.72 308
weighted avg 0.75 0.76 0.75 308
Finally, append a new feature column in the test dataset called Prediction and print the dataset. Code Output  Concluding The Report
|